
| Product | Application | Size | Price | (Price/Library) | Quantity |
|---|---|---|---|---|---|
| SLALOM 1.0 ™ sgRNA Library Synthesis Kit (T7 Transcription) | In vitro Transcription | 10 reactions | $1,240.00 | ($124.00) |
-
+
|
| SLALOM 1.0 ™ sgRNA Library Synthesis Kit (LentiCRISPRv2) | Lentiviral Packaging | 10 reactions | $1,240.00 | ($124.00) |
-
+
|
| SLALOM 1.0 ™ sgRNA Library Synthesis Kit (Mobile CRISPRi) | Bacterial Screening | 10 reactions | $1,240.00 | ($124.00) |
-
+
|
|
Go to Cart
|
PRODUCT INFORMATION
SLALOM 1.0 ™ sgRNA Library Synthesis Kits
USER MANUAL
For a detailed protocol or additional information about the kit, download the complete user manual.
OVERVIEW
SLALOM 1.0™ sgRNA Library Synthesis Kits, S. pyogenes provide a simple and quick method for enzymatically synthesizing high quality sgRNA libraries through a sequence of fast reactions, using the supplied reagents and source of DNA supplied by the user.
The CRISPR Cas9 complex from S. pyogenes can be programmed to bind to a specific DNA sequence by simply altering the first 20 nucleotides of a single guide RNA (sgRNA)¹. Libraries of sgRNA molecules have enabled new techniques including live cell chromatin imaging and functional genomic screening, but the synthesis of custom libraries can often be a bottleneck in these experiments. SLALOM 1.0 ™ sgRNA Library Synthesis Kits permit the synthesis of a custom library in a few hours and without specialized bioinformatic techniques.
SLALOM 1.0™ sgRNA Library Synthesis Kits, S. pyogenes are based on SLALOM (sgRNA Library Assembly by Ligation Onto Magnetic beads)², which works on the principle of eznymatically extracting valid spacer sequences from a source of DNA. These kits were designed to be quick and highly efficent and include optional control DNA and a troubleshooting guide to ensure fast synthesis of custom libraries.
Figure 1: Workflow of SLALOM 1.0 ™ sgRNA Library Synthesis Kits, S. pyogenes.
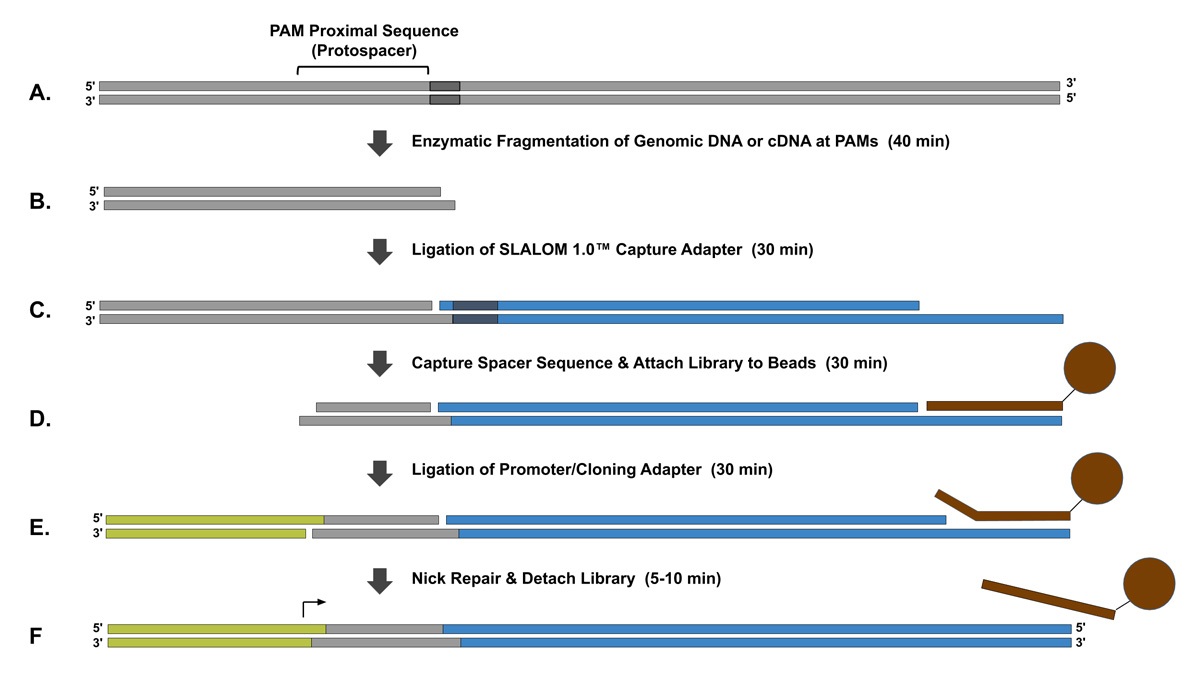
(A). A source of DNA such as Genomic DNA or cDNA is isolated. (B). The DNA source is enzymatically digested at protospacer adjacent motifs (PAMs) to create DNA fragments with valid spacer sequences on each end of the fragments. (C). The SLALOM 1.0™ Capture Adapter is ligated to the ends of the fragments. (D). Using a restriction enzyme that cleaves approximately 20 bp away from its reqconition sequence, the spacer sequences are captured and the remaining fragments are washed away using bead purification. (E). A promoter or cloning adapter is ligated to the spacer sequence. (F). The final library is detached from the beads and nicks in the library are repaired. The library can be used as a template for in vitro transcription or cloned into plasmids depending on the downstream application.
RELATED BLOG POSTS
Genome-wide Screening in Diverse Bacteria with SLALOM and Mobile CRISPRi
Transcriptome-wide Screening in Cell Culture with SLALOM and lentiCRISPRv2
COMPONENTS
The following materials are supplied in these kits:
- SLALOM 1.0 ™ sgRNA adapter
- Promoter/Cloning adapter
- Capture Beads
- Wash Buffer
- C1 Buffer
- C1 Enzyme
- C2 Buffer
- C2 Enzyme
- C2 Enhancer
- L1 Buffer
- L2 Buffer
- L1/L2 Enzyme
- E1 Buffer
- E1 Enzyme
- Control DNA
- User Manual
The following materials are supplied by the user:
- 1-2 μg DNA (per library)
- 1.5 mL microcentrifuge tubes
- Magnetic Rack for 1.5 mL microcentrifuge tubes
References
1. Yates JD, Russell RC, Barton NJ, Yost HJ, Hill JT. A simple and rapid method for enzymatic synthesis of CRISPR-Cas9 sgRNA libraries. Nucleic Acids Res. 2021;49(22):e131.
2. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816-821.
LEGAL
This product is covered by one or more patents owned by Pioneer Biolabs, LLC.
The use of this product may require the buyer to obtain additional third party intellectual property rights for certain applications.
For more information about commercial rights, please contact us at support@pioneerbiolabs.com.
This product is for research use only. This product is not for use in diagnostic procedures.