MICROBIAL CELL ENGINEERING
Engineering microbial cells that produce high amounts of heterologous protein can be difficult.
We are currently developing a genomic screening platform that can be used to test thousands of genomic modifications simultaneously to quickly generate data that can be used as a roadmap for increasing expression of a target protein in a specific cell type.
With this new platfrom companies can achieve faster development times, better bioprocesses, and strains that outcompete other players in the industry.
CRISPR LIBRARY GENERATION
SLALOM 1.0™ sgRNA Library Synthesis Kits, S. pyogenes provide a simple and quick method for enzymatically synthesizing high quality sgRNA libraries through a sequence of fast reactions, using the supplied reagents and source of DNA supplied by the user.
The CRISPR Cas9 complex from S. pyogenes can be programmed to bind to a specific DNA sequence by simply altering the first 20 nucleotides of a single guide RNA (sgRNA)¹. Libraries of sgRNA molecules have enabled new techniques including live cell chromatin imaging and functional genomic screening, but the synthesis of custom libraries can often be a bottleneck in these experiments. SLALOM 1.0 ™ sgRNA Library Synthesis Kits permit the synthesis of a custom library in a few hours and without specialized bioinformatic techniques.
SLALOM 1.0™ sgRNA Library Synthesis Kits, S. pyogenes are based on SLALOM (sgRNA Library Assembly by Ligation Onto Magnetic beads)², which works on the principle of eznymatically extracting valid spacer sequences from a source of DNA. These kits were designed to be quick and highly efficent and include optional control DNA and a troubleshooting guide to ensure fast synthesis of custom libraries.
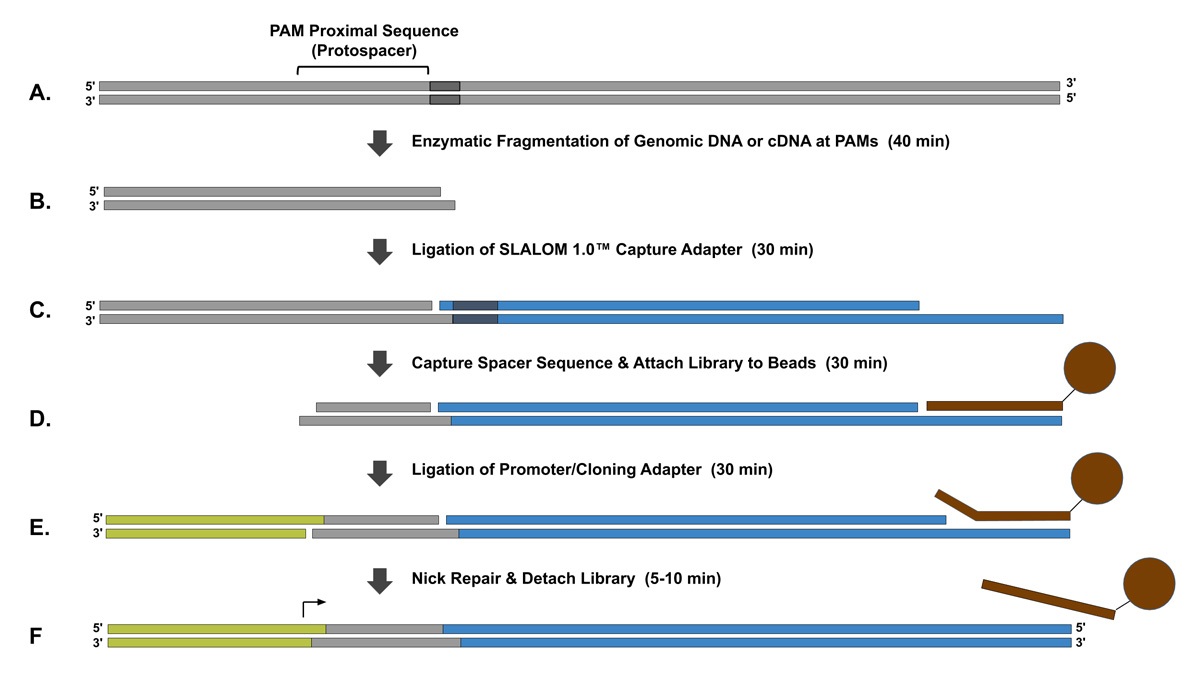
Figure 1: Workflow of SLALOM 1.0 ™ sgRNA Library Synthesis Kits, S. pyogenes.
(A). A source of DNA such as Genomic DNA or cDNA is isolated. (B). The DNA source is enzymatically digested at protospacer adjacent motifs (PAMs) to create DNA fragments with valid spacer sequences on each end of the fragments. (C). The SLALOM 1.0™ Capture Adapter is ligated to the ends of the fragments. (D). Using a restriction enzyme that cleaves approximately 20 bp away from its reqconition sequence, the spacer sequences are captured and the remaining fragments are washed away using bead purification. (E). A promoter or cloning adapter is ligated to the spacer sequence. (F). The final library is detached from the beads and nicks in the library are repaired. The library can be used as a template for in vitro transcription or cloned into plasmids depending on the downstream application.
References
SLALOM 1.0 Capture Adapters ™
Dual functioning adapters make library synthesis fast and efficient.
SLALOM 1.0 ™ uses specially designed adapters that function both as a binding site for a restriction enzyme and as a template for sgRNA transcription. The restriction enzyme that binds to the adapter is able to cleave approximately 20 nucleotides away from it's recognition sequence which makes the adapter useful for capturing spacer sequences from DNA substrates.
This adapter can also be quickly attached and detached from magnetic beads by incubating the beads in the correct solution for a few minutes. The dual functioning adapter in combination with magnetic bead purification makes library synthesis highly efficient, allowing the user to work with small amouts of DNA.
SLALOM 1.0™ sgRNA Library Synthesis Kits
We have created a simple kit that can help you use this technology in your workflow.
SLALOM 1.0™ sgRNA Library Synthesis Kits, S. pyogenes provide a simple and quick method for enzymatically synthesizing high quality sgRNA libraries through a sequence of fast reactions, using the supplied reagents and source of DNA supplied by the user.
